
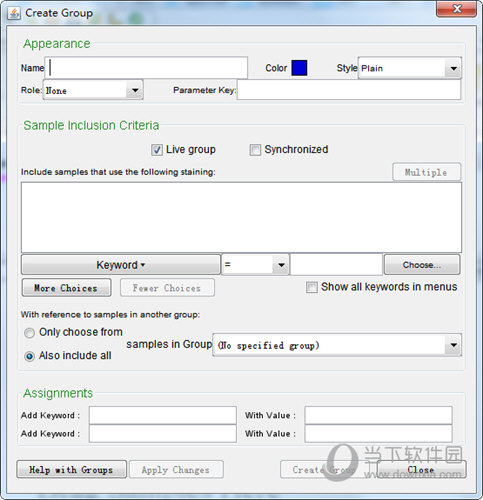
You can also manually drag any samples into any group you’ve created. If you assigned a set of keyword restrictions to the group, the samples that match the keyword restrictions will be present in the group automatically. You can also make “manual” groups in which you just manually drag and drop samples into the group within the workspace.Īfter creating a new group, it will appear within the group panel of the workspace. Note: You don’t have to utilize keywords. This could be a specific staining protocol introduced during acquisition or a keyword combination and could include reference samples in another group. Select the sample inclusion criteria for groups.This will help the user know the purpose of creating a particular group. The choices available are “Test ”, “Replicate”, “Compensation”, “Baseline”, “Gating Level”, “Controls”, and “Instrumentation”.

Choose whether or not to make the Group “Live” (FlowJo will examine all samples subsequently added to the Workspace and add them to a group if they match that group’s criteria) and/or “Synchronized” (FlowJo will automatically apply changes - namely gate adjustments - made in one of the samples of the group to all other samples within that group).Give the Group a name, color, and text style.The basic process for creating a group is to: This window also appears when you double-click on a group in order to edit the criteria for membership. The results on survival were inconclusive (partially due to contamination), but the results on PHB use suggest that variation in PHB accumulation in nodules could contribute to fitness of individual rhizobia long after they are released into the cell.When you click the Create Group button (under the FlowJo tab or in the static toolbar) you are presented with the dialog window below. The experiments compared survival and PHB use among rhizobia isolates with heritable variation in initial PHB supply, incubated in starvation cultures for over a year. The model shows that the levels of PHB accumulated in nodules could potentially support survival functions for many years after symbiosis, based on published metabolic parameters and soil temperature measurements. Part 2 explores the implications of heritable PHB variation for variation in fitness during the free-living stage using modeling and laboratory experiments.
#Flowjo 7.6 1 plus#
The conclusions are based on observations of phenotypic variation in rhizobial PHB per cell measured in the field (soybean and Chamaecrista fasciulata) and in greenhouse plants inoculated with field soil, plus experiments with rhizobia isolates showing high heritability in PHB per cell within soybean nodules. Part 1 provides evidence for wide-ranging, heritable phenotypic variation within natural rhizobia populations in the amount of PHB stored during symbiosis. The paper addresses two gaps in knowledge needed to understand how polyhydroxybutyrate (PHB), a storage compound accumulated by many nitrogen-fixing rhizobia during interactions with legume hosts, contributes to the in the evolution of the legume rhizobia mutualism. The data are being released to meet a publisher's data sharing requirements. This repository contains all analysis scripts and data files for Katherine Muller's first dissertation chapter: Resource acquisition and allocation traits in symbiotic rhizobia with implications for life-history outside of legume hosts.


 0 kommentar(er)
0 kommentar(er)
